
8. Data extraction and coding
This session will cover how to extract data from studies, both in terms of descriptive information (meta-data) and quantitative (or qualitative) study findings. It will also describe coding of studies, which is used in systematic mapping. This involves populating an interactive database with predefined codes that allow users to rapidly search for relevant studies based on a suite of information about the study setting and methods. This session will involve a practical exercise where attendees will plan and undertake basic coding and meta-data extraction.
Learning objectives:
- To appreciate the importance of careful planning before undertaking data extraction
- To understand the difference between meta-data extraction and quantitative/qualitative data extraction
- To understand how to undertake coding of studies
- To appreciate how to plan data extraction in a repeatable, reliable manner
- To understand the importance of consistency checking/double checking of data extraction activities
To begin, watch the following presentation:
You can find the lecture handouts here.
Next, read the seventh chapter of the guidance from the Collaboration for Environmental Evidence here. We'll revisit sections of the CEE Guidance throughout the course. It focuses on environmental and conservation topics, but is subject agnostic, meaning that it's useful for other disciplines, too. Feel free to focus instead on the Cochrane Guidance if you work with healthcare topics.
Practical exercise
In this exercise, you will gain experience of another free systematic review management software, SysRev.com. This hypotheical expample examines the impact of marine microplastics on biodiversity. Your task is to extract data and code studies according to a predefined schema (set of variables). Follow the instructions below. Thanks to the SysRev team for helping to develop this exercise.
You have been recruited into a team conducting a systematic map on evidence of the effects of marine plastics on organism health. The map is progressing well and is now at the meta-data extraction and study coding stage. At this point, you have been given 5 articles for which meta-data extraction and coding must be conducted. The articles cover a range of systems and organisms, some of which includes human food species.
For this review, you will be using the web application SysRev.com. For an overview of SysRev, the developers have put together this demo.
Click on the following link to set up an account and join the review: https://sysrev.com/register/21d40d5e9c45.
Once you have joined the review, you will see the platform has three tabs: 'Overview' (which displays high-level graphics on the progress of the review), 'Articles' (where you can filter and see the list of articles in the project), and ‘Review’ (where you can extract meta-data and code studies:
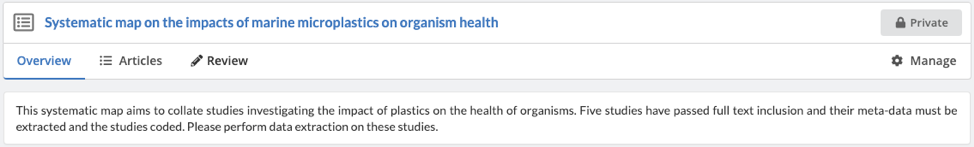
As a review member, you will be able to extract meta-data and code articles but not edit the settings of the review ('Manage').
To begin reviewing articles, click on the 'Review' tab and Sysrev will automatically display the first of five articles that have been uploaded. By default, Sysrev shows the 'Abstract' view. Click on 'PDF' to view the full text. In order to navigate within a given PDF, use the 'Previous' and 'Next' buttons, shown below.
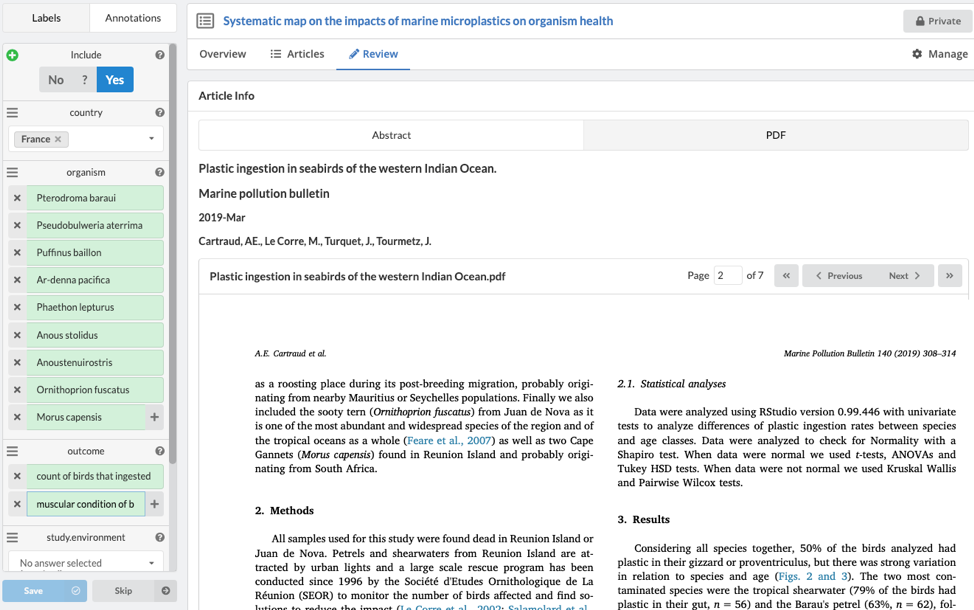
In the left-hand panel, you will see the 'Label' prompts – that is, the meta-data and coding to be extracted from each article.
Take some time to read the objectives, methods, and results of each paper and fill in the Label prompts as best you can. Sysrev facilitates three types of labels: Boolean (True/False), Categorical (pre-defined choices), and String (free text responses). In combination, the label prompts give the reviewer flexibility in which (and how much) data is extracted.
When extracting data, you can select multiple values for bot drop-down labels (coding) and free text labels (add new lines by using the '+' button for each variable). You will need to complete all 'required' labels, as well as designate ‘Yes’ for inclusion before saving and reviewing the next article.
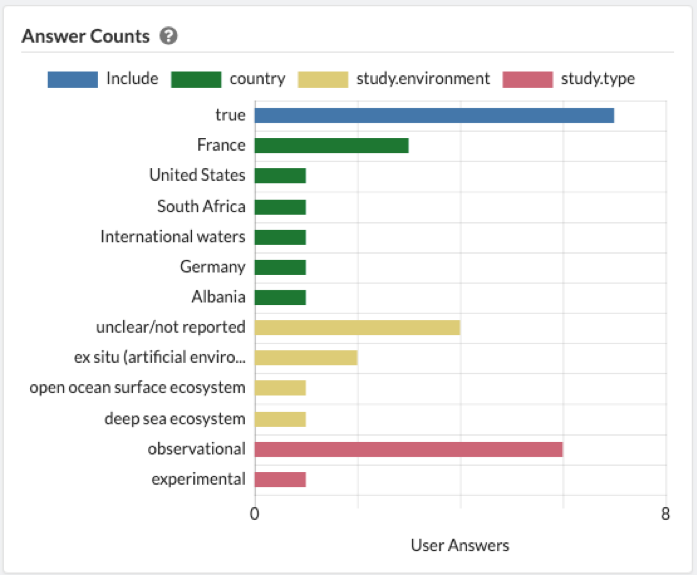
Once an article has been saved, the data is automatically logged in both the 'Overview' and 'Articles' tab.
The 'Articles' tab shows which reviewers have reviewed each article. By clicking on a specific article, you can see which labels were ascribed by each reviewer. If you wish to alter the extracted data, simply click ''Change Labels' to be brought back to the article review screen.
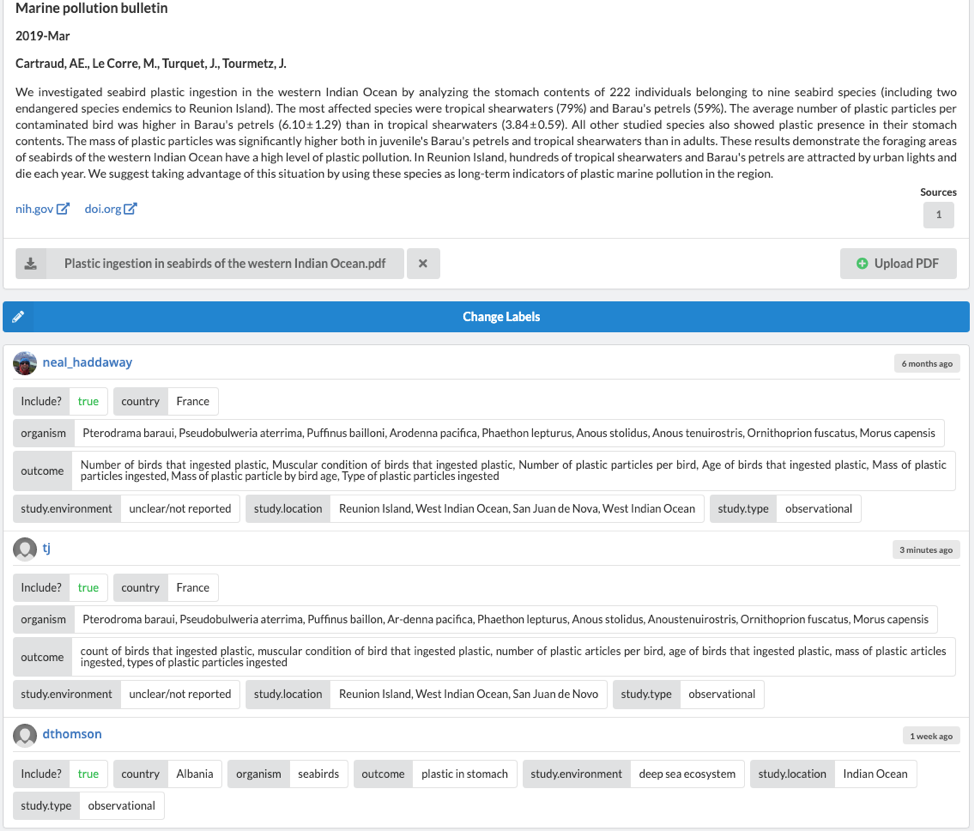
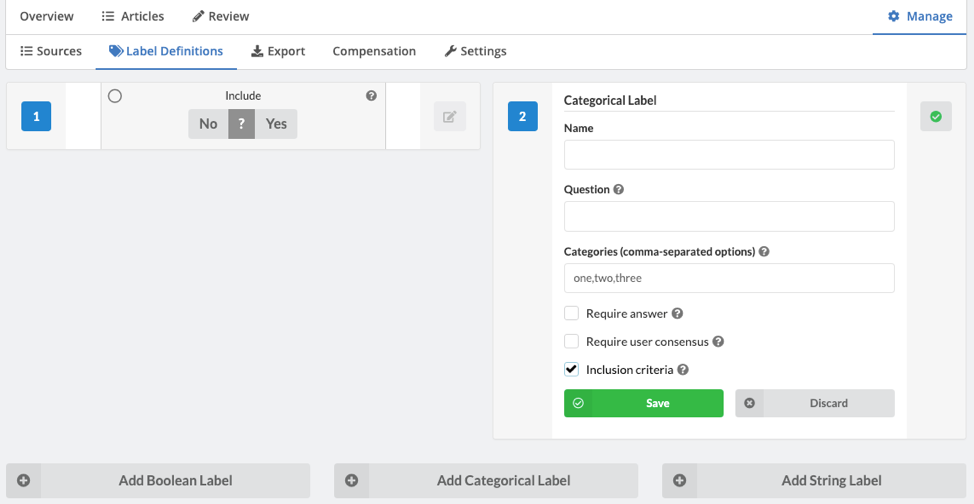
If you'd like to see how the data extraction was set up, create your own new review and proceed to 'Manage>Label Definitions'. You can now create as many boolean, categorial, and string 'Labels' as required by your project. For boolean and categorical labels, you can also designate any label as "inclusion criteria" which stipulates a specific answer as being required for inclusion. This is a useful way to clarify inclusion requirements to your reviewers.
Once you have finished the review, you can download the map database by clicking 'Export', then either 'Group Answers' (one line entry per article, all answers consolidated across reviewers) or 'User Answer' (one line entry per reviewer per article), and then click 'Generate'. The CSV file is then ready for download.
So now you have experience of how to extract data and code studies, let's spend some time thinking about how we critically appraise studies for their validity.
Move to the next module!